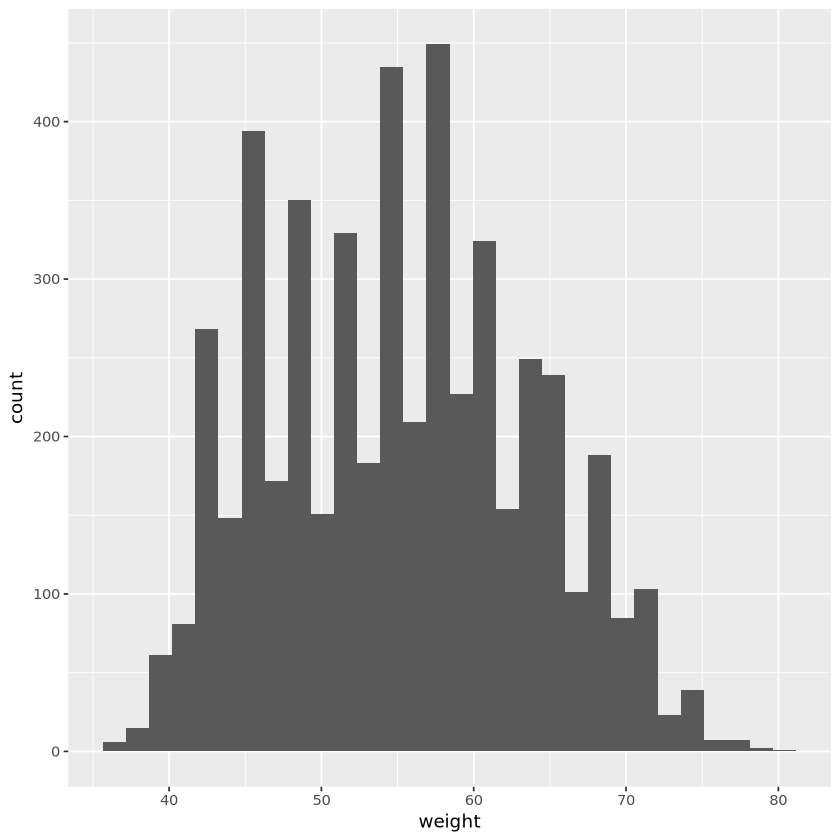
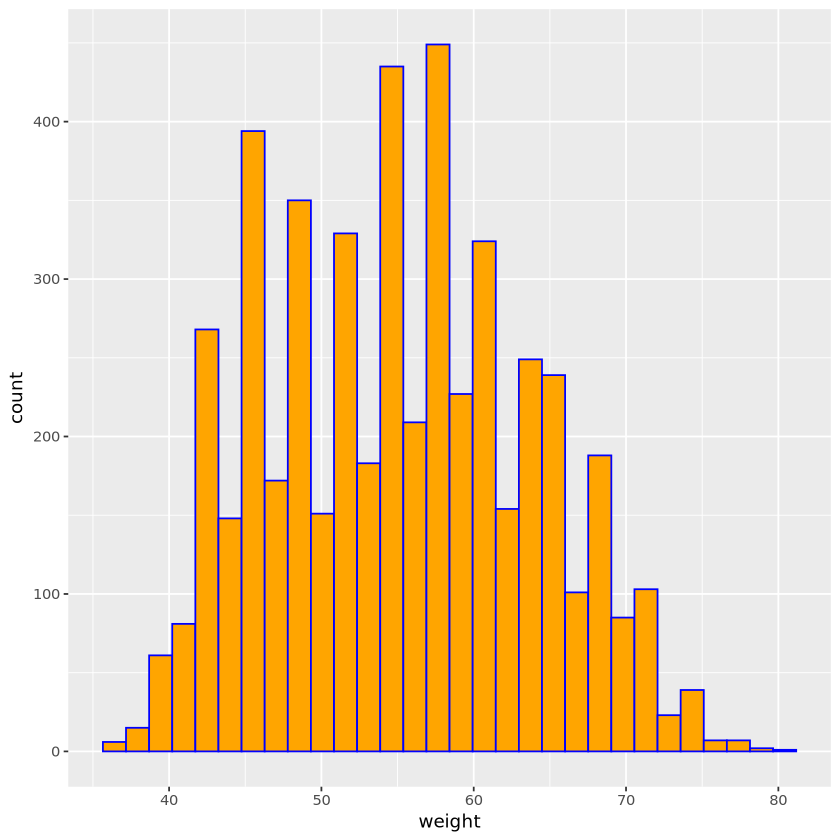
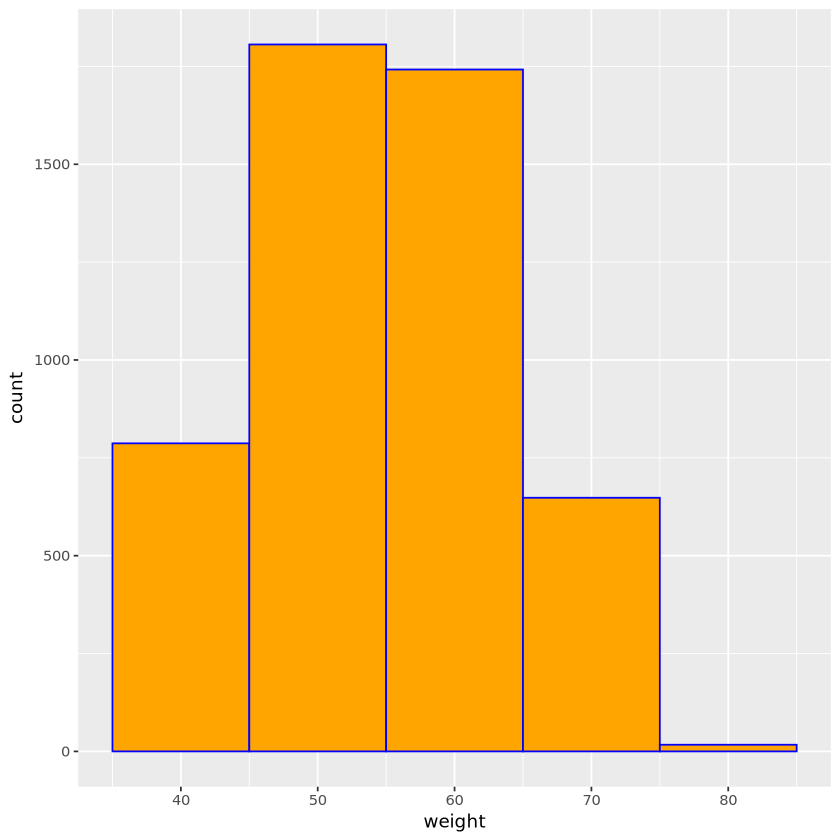
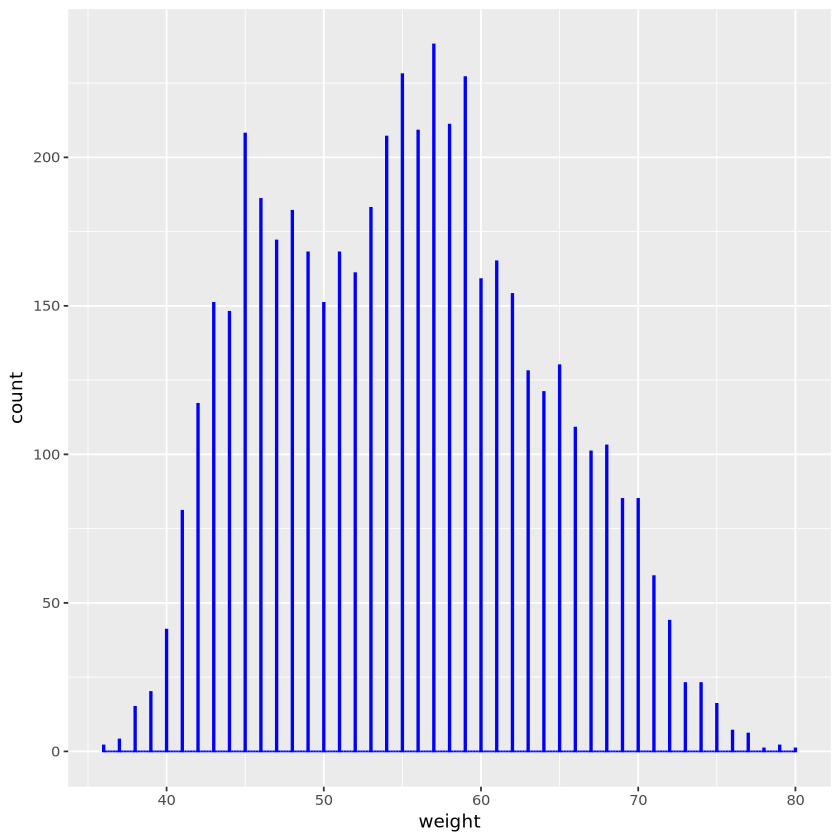
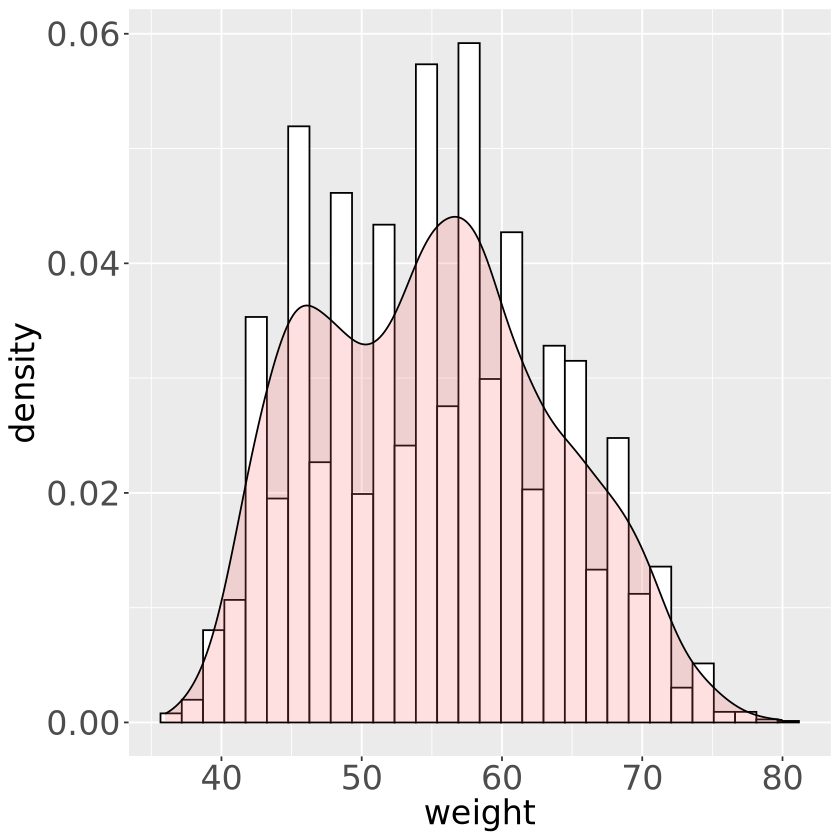
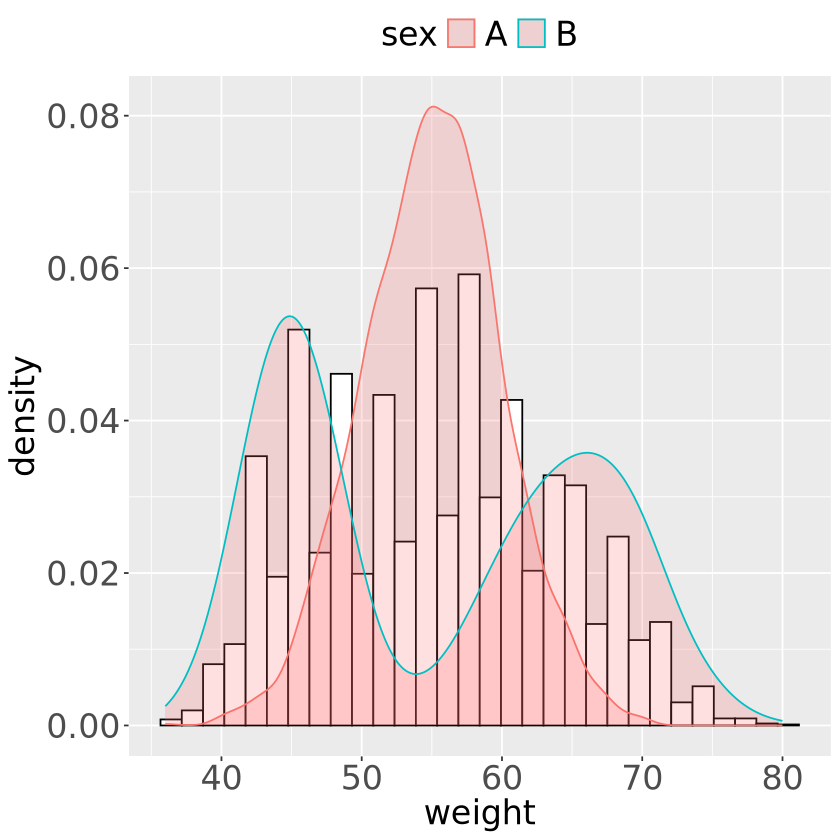
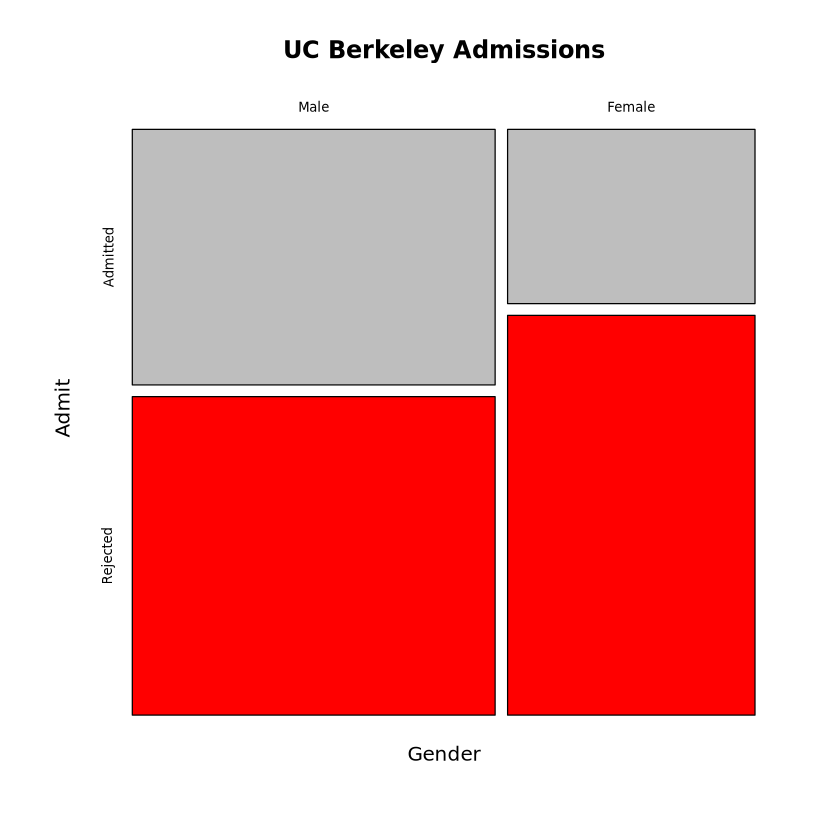
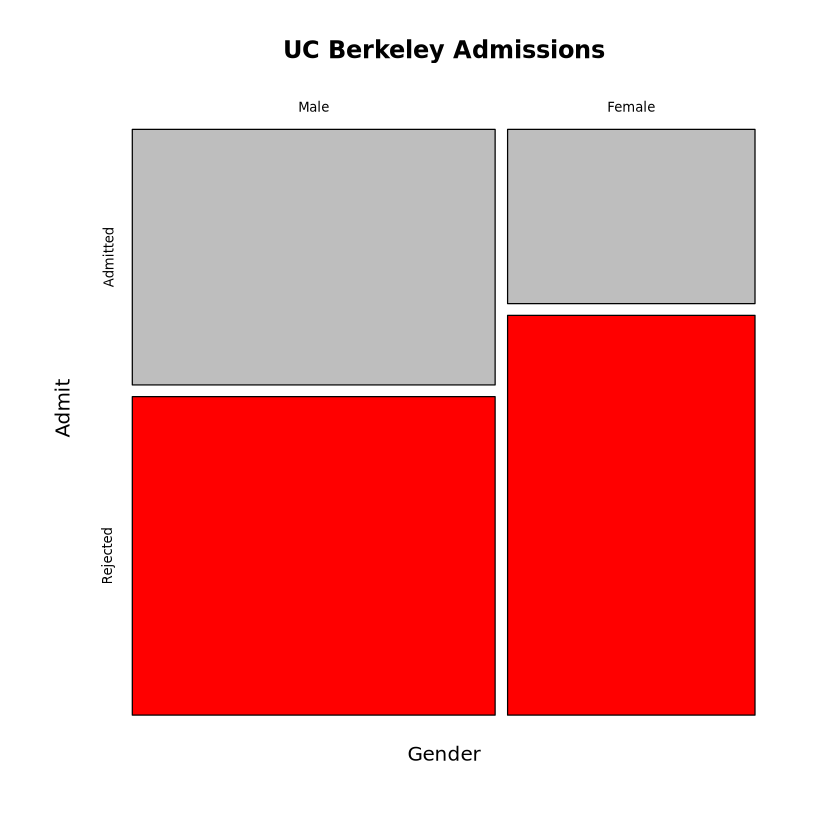
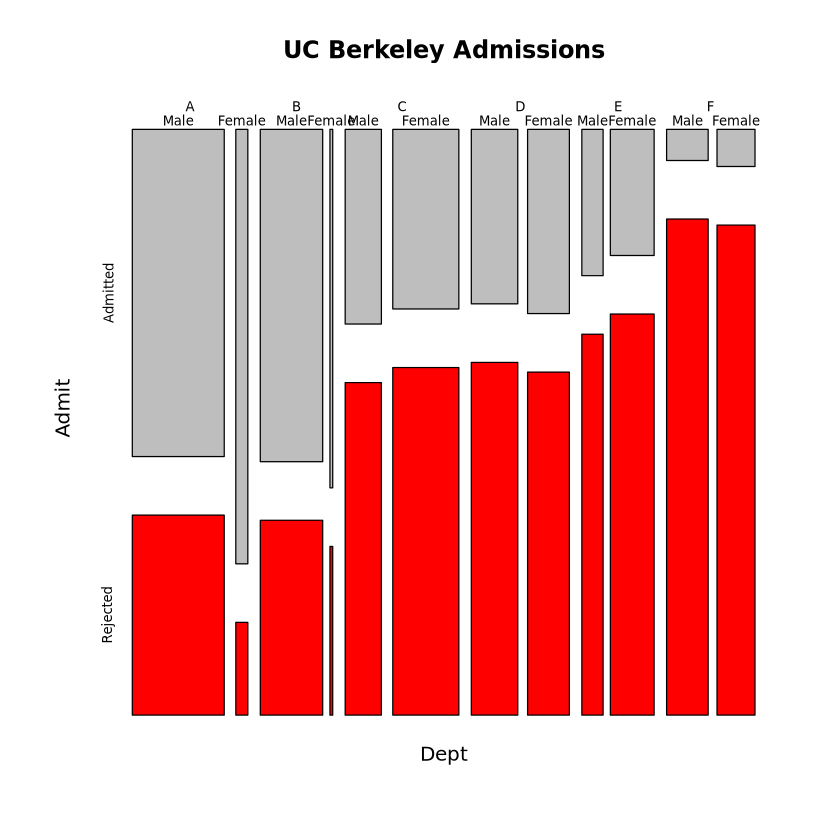
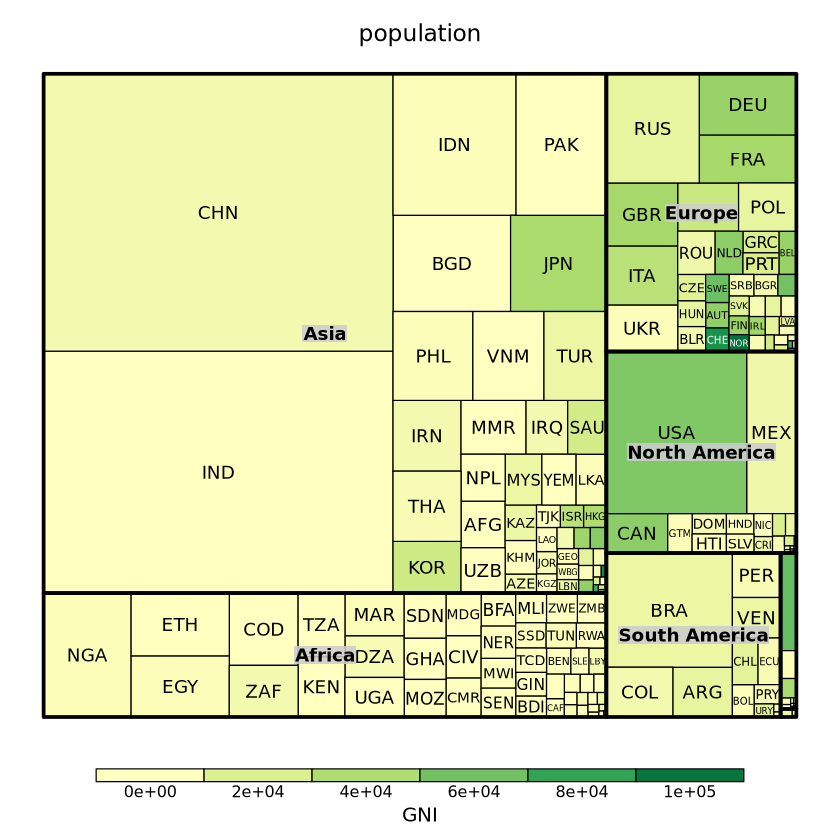
set.seed(11)
rbinom(2500, 1, 0.5) -> ipp
df <- data.frame(
sex=factor(rep(c("A", "B"), each=2500)), weight=round(c(rnorm(2500, mean=55, sd=5),
ipp* rnorm(2500, mean=65, sd=5) + (1-ipp)*rnorm(2500, mean=45, sd=3) ) ))
head(df) ; tail(df)| sex | weight | |
|---|---|---|
| <fct> | <dbl> | |
| 1 | A | 56 |
| 2 | A | 64 |
| 3 | A | 57 |
| 4 | A | 48 |
| 5 | A | 55 |
| 6 | A | 57 |
| sex | weight | |
|---|---|---|
| <fct> | <dbl> | |
| 4995 | B | 65 |
| 4996 | B | 45 |
| 4997 | B | 68 |
| 4998 | B | 66 |
| 4999 | B | 47 |
| 5000 | B | 63 |